Bioreactor control breakthroughs: Biopharma industry turns to advanced measurements, simulation and modeling
The biopharmaceutical industry is one of the fastest growing and most sophisticated of the 21st century. Biologically produced therapies play a significant role in improving life expectancy and quality of life around the globe. Presently, biopharma accounts for about 20% of the overall pharma market, generating global annual revenues over $230 billion USD. At the center of these therapies is a biochemical process in a bioreactor, which provides a controlled environment for optimal cell growth and/or product formation.
Bioreactor batches for new biologics may take 10 or more days to complete, and can be worth $10 million USD or more. The loss in production time and profit from a bad batch is enormous. Meanwhile, the cellular kinetics and the response complexity and dynamics are more challenging than most any other process. It's unreasonable for operators or process engineers to be able to diagnose and correct problems after a few weeks of analysis, let alone during the production of a given batch.1
The best process control is essential to repeatable batch production with maximum yield. Adverse conditions must be recognized and corrected before a bad batch becomes inevitable, and the biopharma industry is increasingly turning to new, advanced measurement techniques as well as modeling and simulation tools such as digital twins to produce consistent product.
Advances in measurement
There are many inline measurements offered by the relatively inexpensive use of improved probes for dissolved oxygen (DO), dissolved carbon dioxide and biomass. Inline or online biomass measurement techniques based on turbidity, dielectric spectroscopy and near infrared spectroscopy are available. Then there's the at-line mass spectrometer, the extremely powerful Bioprofile Flex analyzer and fast protein liquid chromatographs. The breakthroughs involve the ability to provide inferential measurements and consequential process control improvements. This supports analysis of beneficial and adverse operating conditions and concentrations leading to cell health, cell growth rate, cell death rate, and production rate indications and predictions.
DO is measured by an inline electrode through the amperometric principle of an electrochemical sensor. Oxygen from the process crosses the sensor membrane. This membrane is molecule selectable, which means only oxygen can cross to the cathode. DO is important to measure because cells quickly shut down and die from low DO content, which is modeled by the limitation parameter in the Michaelis-Menten kinetics equation. Not as well recognized is that a cell can also suffer from very high DO, which is defined using the inhibition parameter in the equation. Tight oxygen control is essential for accurate computation of oxygen uptake rate (OUR), which is used in turn to estimate cell growth rate, product formation rate and gas mass transfer coefficients (KLa).
Cell metabolism results in a dissolved carbon dioxide production rate (CPR). For biologics, carbon dioxide is manipulated for pH control, lowering the media pH by an increased concentration of carbonic acid. Dissolved carbon dioxide (DCO2) buildup that's detrimental to cell health often occurs in large bioreactors at low gas flow rates and with poor pH control often associated with unnecessary crossings of the split range point. DCO2 is measured by an inline Potentiometric Severinghaus Electrode. The Severinghaus Electrode uses a bicarbonate buffer system in contact with the measured media through a selective CO2 permeable membrane.
Turbidity provides an inferential measurement of total cell mass including intracellular product. Turbidity measurements are based on Beer-Lambert law, an optical principle taking advantage of the interaction of light with undissolved (suspended) particles. Turbidity is determined by measuring the intensity of the scattered light at a particular angle or by the loss of light intensity in transmission.
Dielectric spectroscopy provides a measurement of viable cell mass, which by subtraction from total cell mass gives a measure of dead cell mass. The rate of change of dead cell mass is death or lysis rate. The rate of change of turbidity is an indication of cell growth rate and product formation rate assisted by OUR and oxygen yield factors for growth, maintenance and product formation. The associated CPR improves the computation of the product formation rate. Measurement of glucose and glutamine concentrations with yield factors can also improve inferential measurements.
Normal healthy animal cells have a relatively constant spherical cell size. As cells go through a natural death phase termed apoptosis, their shapes become irregular and they shrink. Fogale dielectric spectroscopy uses an inline probe to make random measurements between 0.1 and 15 MHz and a non-linear least squares fit to generate the entire frequency spectrum. The resulting peaks and beta dispersion areas in permittivity at various frequencies can be used to infer cell volume, size, and homogeneity and membrane integrity.
The concentration measurement of energy sources, nutrients and toxic byproducts enables more optimum scheduling of feeds in the batch sequence and even fed-batch closed loop control of key concentrations. For example, a buildup of glucose and glutamine can cause excessive production of lactate and ammonia byproducts, respectively, which are detrimental to cell health and can inhibit cell growth. It's possible to model such effects using the inhibition parameters for these components in Michaelis-Menten equations.
Ammonia production also necessitates the addition of sodium bicarbonate for pH control that causes high cell osmotic pressure. A glucose and glutamine concentration that's too low can reduce cell growth as well, and is incorporated in the limitation parameters for these components in Michaelis-Menten equations. Precursors to cell apoptosis, such as metabolized amino acid and nicotinamide adenine dinucleotide+hydrogen (NAD+H), can also be measured. Near-Infrared spectroscopy (NIR) provides measurements of chemical groups from the absorption bands in the mid infrared spectrum that are particularly strong. This can be done without sample preparation by inserting a fiber-optic probe (e.g., Ingold port-adapted probe with 4-mm path length from Mettler-Toledo) into the process. Fiber-optic cables can be used to send the probe signal to a remote analyzer (e.g., Aspectrics encoded photometric NIR).
Measuring oxygen, carbon dioxide and nitrogen concentrations in the vent gas flow of a bioreactor enables computation of the OUR, CPR and respiration quotient (RQ), which can be an indicator of abnormal operating conditions. Meanwhile, the ability to measure concentrations of all components in the media, including glucose, glutamine, glutamate, lactate, ammonia, sodium and potassium with a resolution of 0.01 g/L plus cell density, cell diameter, cell viability and osmolality, provides a nearly complete picture of cell health.
Digital twins accelerate discovery
Often, biopharmaceutical production process are designed based on laboratory benchtop and pilot plant runs. Even here, the window of opportunity is narrow since batch cycle times are extremely long—especially for new biologics (e.g., 10 days for mammalian cell bioreactors), and the time to market must be minimized to make the most out of patent protection for high-value products.
One way to address these time limitations is with the effective use of a digital twin of the production process. Experimentation was once largely limited to early phases of R&D, but recent breakthroughs in kinetic modeling enable model parameters to be readily set so that the batch profiles in the digital twin match closely those in the lab and plant. Exploration with first principle models, data analytics, inferential measurements and control strategies in the digital twin offers innovation to improve R&D and knowledge to sustain batch performance in production.
Such digital twins enable rapid and free experimentation with acceleration factors greater than 500 times the actual process, meaning that a 10-day batch can be simulated and analyzed in 30 minutes. Digital twin systems allow users to quickly tour the full cycle of discovery, from creating hypotheses to performing experiments to analyzing and interpreting results.
The speedup of the process model can be much faster than the real-time multiplier for the control system. The process model speedup is the product of speedup factors for kinetics and the material and energy balances. If the kinetic speedup factor is 10 and the material and energy balance factor is 50, the process model speedup is 500. If the control system speedup is 10, the response times of the instrumentation can be reduced by a factor of 50 to keep the PID tuning about the same. Maximum valve, pump and measurement flows are increased by the kinetic speedup factor to match the increase in growth and formation rates.
Increasingly, researchers are realizing the value of improved capability and flexibility of a small DCS to run batches automatically in a lab or pilot plant, greatly reducing cycle time and nonintentional variability. The transition from lab to pilot plant to production plant of the actual control system and digital twin is seamless and fast, greatly reducing the validation burden.
[javascriptSnippet]
Equations have been found that enable the user to readily set parameters that provide robust, high-fidelity kinetics for the effects of temperature, pH and concentrations on cell growth rate and production rate. Convenient cardinal equations can match the shape and peaks of the nonsymmetrical curves of cell growth and product formation rates plotted versus temperature and pH by three readily obtainable parameters.
In this model, the optimum temperature or pH for growth and production phases is simply the temperature or pH setpoint including any shifts for batch phases. The minimum and maximum temperatures complete the parameter settings. The model parameter list is minimal and easily configured based on biological observations.
This is a tremendous advancement to traditional uses of Arrhenius equations for temperature and Villadsen-Nielsen equations for pH that required parameters, not readily available, set with a precision to the sixth or seventh decimal place. For concentrations, a generalized Michaelis-Menten equation, shown to be useful for modeling intracellular dynamics, can model the limitation and inhibition effects of concentrations. The equations provide a link between macroscopic and microscopic kinetic pathways. If the limiting or inhibition effect is negligible or needs to be temporarily removed, the limitation and inhibition parameter is simply set to 0 g/L and 100 g/L, respectively. The biological significance and ease of setting parameters is particularly important for the equations presented.
These revolutionary equations enable the same generalized kinetic model to be used for all types of cells. Previously, yeast cells (e.g., ethanol production), fungal cells (e.g., antibiotic production), bacterial cells (e.g., simple proteins), and mammalian cells (e.g., complex proteins) had specialized equations developed that did not generally carry over to different cells and products.
Dynamic first principal models in the literature were primarily done by universities and were for batch processes without control system dynamics. Controllers and strategies were emulated and consequently disparate from actual capability in a modern control system. The dynamic models in a digital twin can operate in the continuous as well as the batch mode with measurement and final control element dynamics and the exact duplicate of controllers, historian, and operator interface in the lab, pilot plant or production unit.
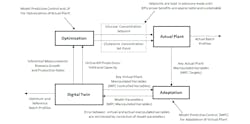
Figure 1: Nonintrusive automated adaptation of model parameters improves fidelity by matching manipulated variables (e.g., PID outputs) in digital twin to those in actual plant with potential future optimization based on improvement in KPIs.
Enter model predictive control
The dynamic model can be adapted online to match the plant by an innovative use of model predictive control (MPC). MPC targets and controlled variables are the actual plant and digital twin manipulated variables (e.g., flows), respectively (Figure 1). The figure illustrates how to adapt a bioreactor model to provide more accurate inferential measurements of biomass growth and production rates that are slopes of the batch profiles for cell and product concentration.
The manipulated variables of the MPC to automate model adaptation are model parameters (e.g., kinetic parameters) that affect these profiles. The MPC can be identified and tested without the digital twin connected to the actual plant. The MPC development for adaptation is much faster than an actual plant MPC. This independence enables extensive adaptation and gaining associated knowledge of process relationships without any disruption to the actual plant. The adaptation is done without affecting the actual plant since the plant’s manipulated variables are being read, and aren't being changed by the digital twin. It's critical that the digital twin have the same setpoints and tuning settings as the actual plant, and the digital twin be started with controller outputs initialized to match the actual plant.
The optimized setpoints from MPC with inferential measurements of growth and production rate and are done in an advisory mode not affecting the actual plant. Not shown in Figure 1 is that an MPC is run in automatic on another digital twin to generate and study the optimized setpoints. The setpoints are only eventually used to automatically optimize the plant if they prove more accurate, beneficial, and reliable per KPIs than an MPC with inferential measurements computed from online and at-line analyzers.
Optimization opportunities
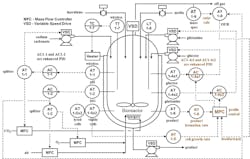
Figure 2: Optimization of cell growth and product formation rates by manipulation of glucose and glutamine concentrations with a preemptive correction based on oxygen uptake rate (OUR).
There are many opportunities for bioreactor optimization through additional control loops (e.g., glucose and glutamine concentration control) using an enhanced PID to better deal with measurement delays² and the addition of a block to compute the rate of change and future value of a concentration measurement.³
PID is the best type of controller in loops where derivative action or a fast response is important, and is best able to deal with the load disturbances on the process inputs. MPC is better able than PID to handle interactions, constraints, disturbance variables (feedforwards), and complex dynamics. MPC is also less sensitive to resolution limits and noise, and can be optimized by an integrated linear program (LP). The best solution often involves PID loops for tight control of operating conditions and MPC to optimize PID setpoints.
MPC can optimize mammalian cell bioreactors by controlling cell growth rate and product formation rate at the setpoints as the batch progresses by the manipulation of glucose and glutamine concentrations. The growth rate is the slope of the batch cell concentration profile and product formation rate is the slope of the product concentration profile. Online and at-line analyzers provide these concentrations directly or by inferential measurements, or the first principle models of a digital twin. The demand for glucose and glutamine changes drastically as cells move through their growth phases (e.g., pre-exponential, exponential and stationary) and into product formation. An OUR computed by analyzing off-gas oxygen content and manipulated air flow and supplemental oxygen flow rates can provide a feedforward signal to provide some anticipation of changes in cell growth rate. Figure 2 shows an MPC application providing this optimization, where a future value block is used to compute cell growth and product formation rates.
All told, much greater bioreactor performance capability and reliability is possible by much better R&D, process design, operations, and control systems facilitated by breakthroughs in easier and more effective use of instruments, digital twins and first principle models.
References
- McMillan, Gregory K., “At the IIoT Crossroads,” Control, Feb. '19, p 36
- McMillan, Gregory K., “Wireless – Overcoming challenges of PID control & analyzer applications,” InTech
- McMillan, Gregory K., “Future PV values are the future,” ControlGlobal.com.